Python Segmentation API
[7]:
# must run in an environment where polarityJaM is installed
from polarityjam import Plotter
from polarityjam import RuntimeParameter, PlotParameter, ImageParameter
from polarityjam import PolarityJamLogger
from polarityjam import load_segmenter
from polarityjam import SegmentationMode
from polarityjam import SegmentationParameter
from polarityjam.utils.io import read_image
from pathlib import Path
[8]:
# Setup a logger to only report WARNINGS, Put "INFO" or "DEBUG" to get more information
plog = PolarityJamLogger("WARNING")
Setup your data
We expect you to be already familiar with basic usage of polarityjam. Hence we will shorten necessary data preparation steps. Please look at the Python API section of the documentation for more information.
[9]:
### ADAPT ME ###
path_root = Path("")
input_file = path_root.joinpath("data/golgi_nuclei/set_2/060721_EGM2_18dyn_02.tif")
output_path = path_root.joinpath("data/polarityjam_out/")
output_file_prefix = "060721_EGM2_18dyn_02"
### ADAPT ME ###
img = read_image(input_file)
# Parameters
params_image = ImageParameter()
params_image.channel_organelle = 0 # golgi channel
params_image.channel_nucleus = 2
params_image.channel_junction = 3
params_image.channel_expression_marker = 3
params_image.pixel_to_micron_ratio = 2.4089
# Parameters
params_runtime = RuntimeParameter()
params_plot = PlotParameter()
# plotter object
plotter = Plotter(params_plot)
Several segmentation algorithms in parallel
Polarity-JaM currently supports three segmentation algorithms out of the box.
Here, we show how easy it is to switch from one to another and how to compare performance.
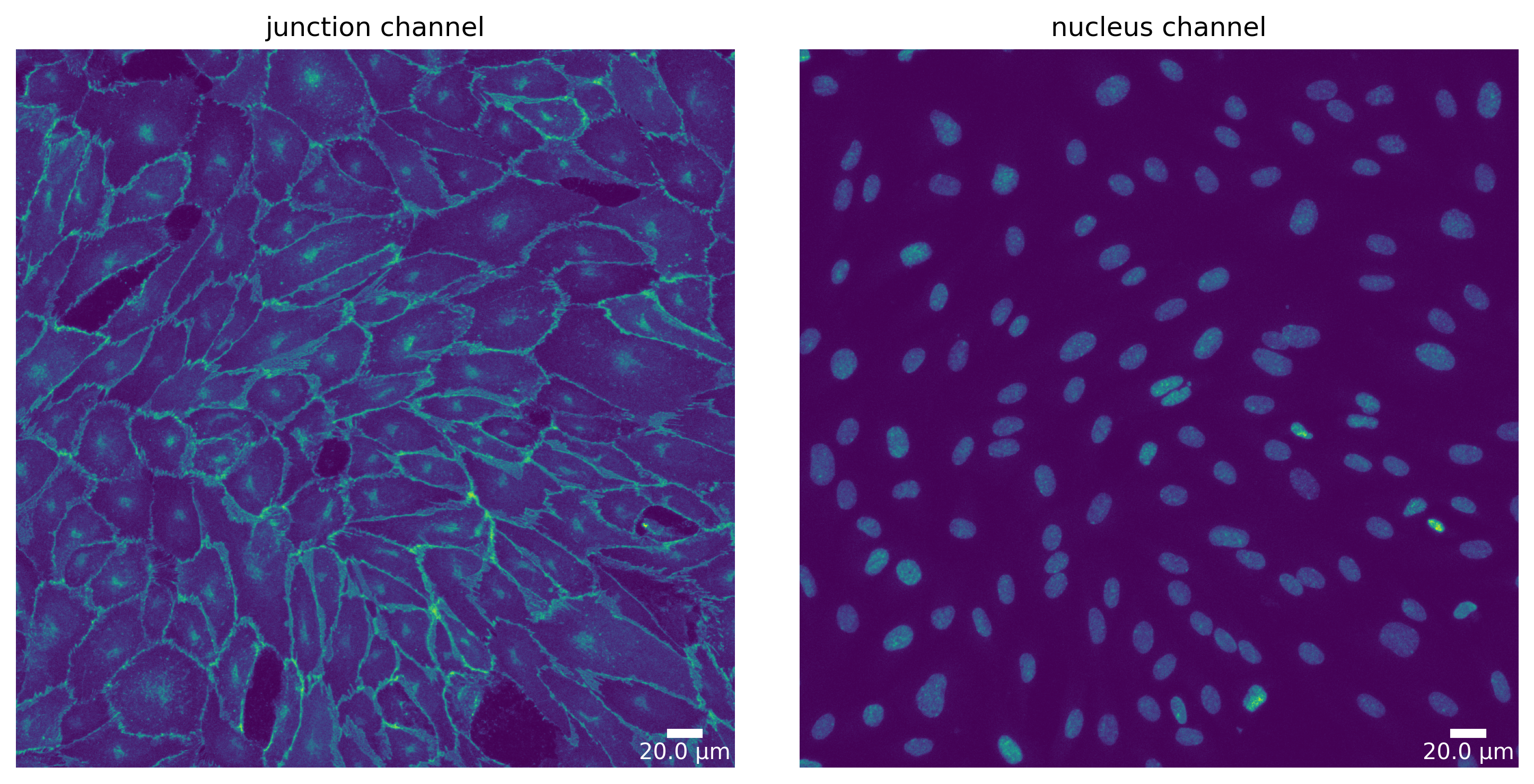
Step 1 - Prepare your image
[10]:
# Cellpose - switch off mask loading (if exists) and saving
cp_seg_p = SegmentationParameter(params_runtime.segmentation_algorithm)
cp_seg_p.use_given_mask = False
cp_seg_p.store_segmentation = False
cp_seg, _ = load_segmenter(params_runtime, cp_seg_p)
cp_seg_prep, cp_seg_prep_p = cp_seg.prepare(img, params_image)
plotter.plot_channels(cp_seg_prep, cp_seg_prep_p, output_path, input_file);
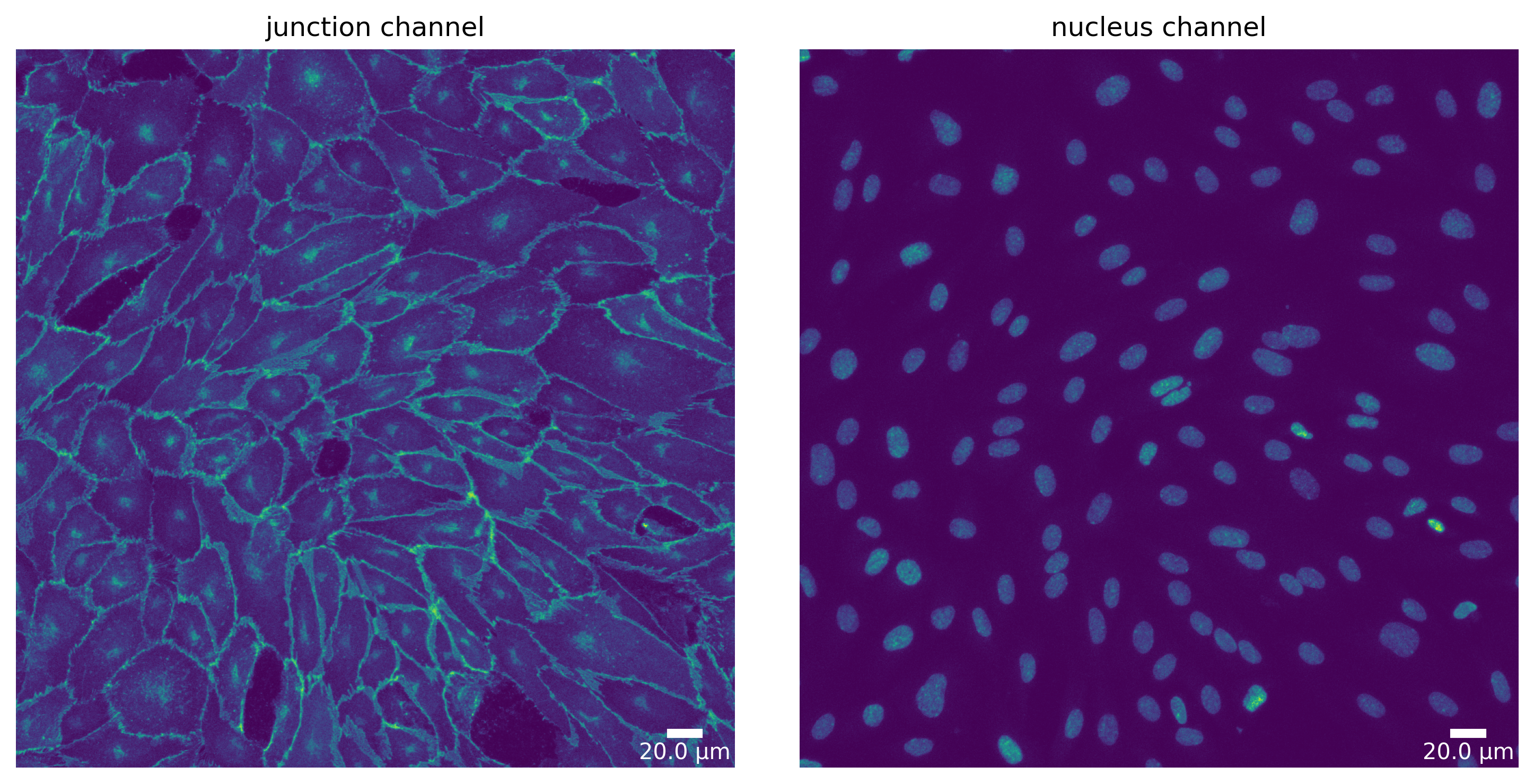
[11]:
# Deepcell - default parameters
params_runtime.segmentation_algorithm = "DeepCellSegmenter"
dc_seg, _ = load_segmenter(params_runtime)
dc_seg_prep, dc_seg_prep_p = dc_seg.prepare(img, params_image)
plotter.plot_channels(dc_seg_prep, dc_seg_prep_p, output_path, input_file);
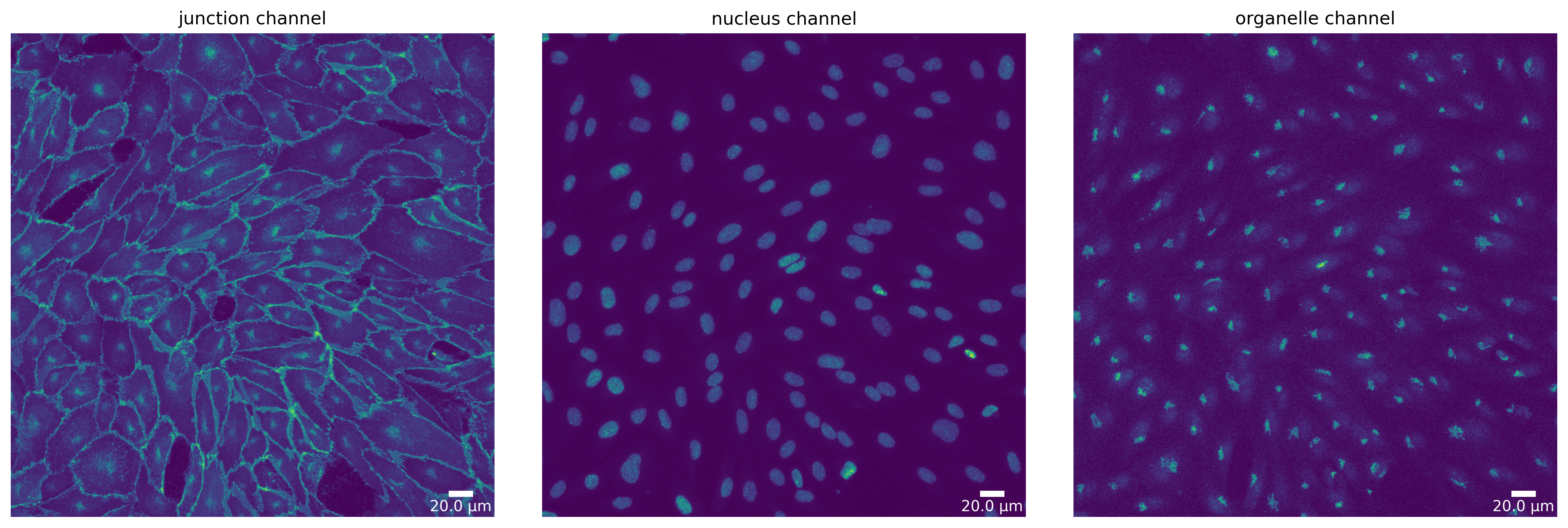
[12]:
# microSam - default parameters
params_runtime.segmentation_algorithm = "MicrosamSegmenter"
ms_seg, _ = load_segmenter(params_runtime)
ms_seg_prep, ms_seg_prep_p = ms_seg.prepare(img, params_image)
plotter.plot_channels(ms_seg_prep, ms_seg_prep_p, output_path, input_file);
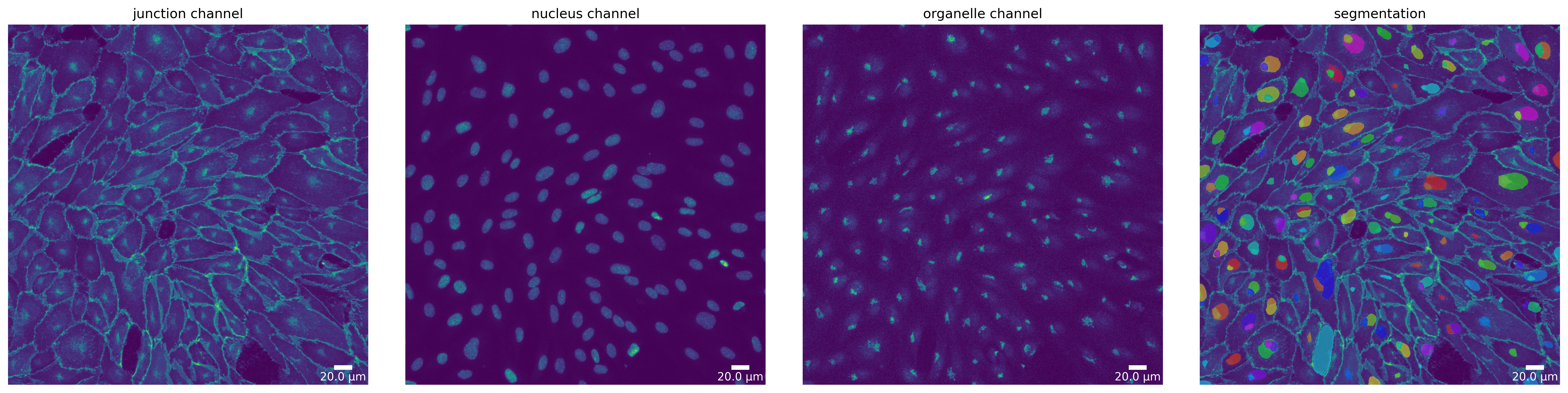
Step 2 - Segment your image
[13]:
# Cellpose
cp_mask = cp_seg.segment(cp_seg_prep)
plotter.plot_mask(cp_mask, cp_seg_prep, cp_seg_prep_p, output_path, output_file_prefix);
cellprob_threshold and dist_threshold are being deprecated in a future release, use mask_threshold instead
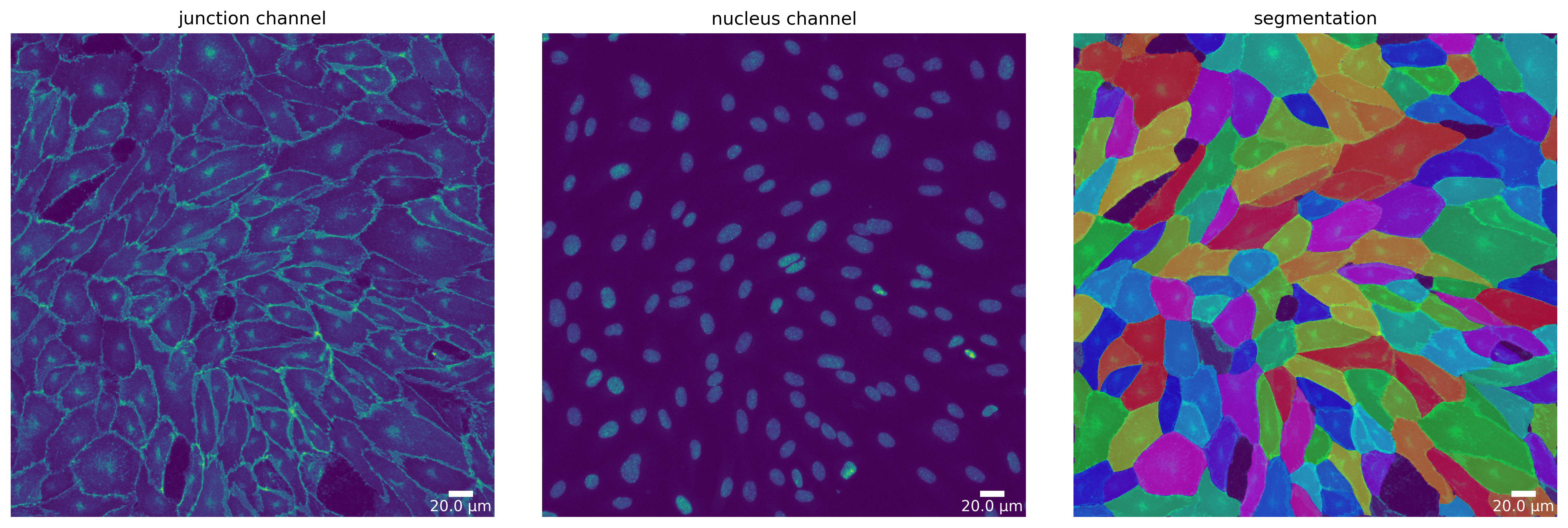
[14]:
# Deepcell
dc_mask = dc_seg.segment(dc_seg_prep)
plotter.plot_mask(dc_mask, dc_seg_prep, dc_seg_prep_p, output_path, output_file_prefix);
17:28:47 INFO
Solution credits:
Noah F. Greenwald and Geneva Miller and Erick Moen and Alex Kong and Adam Kagel and Thomas Dougherty and Christine Camacho Fullaway and Brianna J. McIntosh and Ke Xuan Leow and Morgan Sarah Schwartz and Cole Pavelchek and Sunny Cui and Isabella Camplisson and Omer Bar-Tal and Jaiveer Singh and Mara Fong and Gautam Chaudhry and Zion Abraham and Jackson Moseley and Shiri Warshawsky and Erin Soon and Shirley Greenbaum and Tyler Risom and Travis Hollmann and Sean C. Bendall and Leeat Keren and William Graf and Michael Angelo and David Van Valen; Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning (DOI: https://doi.org/10.1038/s41587-021-01094-0)
17:28:49 INFO ~ Starting DeepCell-predict
17:28:55 INFO ~ 2023-10-27 17:28:55.024477: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX AVX2
17:28:55 INFO ~ To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
17:28:55 INFO ~ 2023-10-27 17:28:55.563949: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 21662 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3090, pci bus id: 0000:80:00.0, compute capability: 8.6
17:29:03 INFO ~ WARNING:tensorflow:No training configuration found in save file, so the model was *not* compiled. Compile it manually.
17:29:03 INFO ~ Image resolution the network was trained on: 0.5 microns per pixel
17:29:06 INFO ~ 2023-10-27 17:29:06.092813: I tensorflow/stream_executor/cuda/cuda_dnn.cc:368] Loaded cuDNN version 8800
17:29:07 INFO ~ 2023-10-27 17:29:07.070662: E tensorflow/core/platform/windows/subprocess.cc:287] Call to CreateProcess failed. Error code: 2
17:29:07 INFO ~ 2023-10-27 17:29:07.071852: E tensorflow/core/platform/windows/subprocess.cc:287] Call to CreateProcess failed. Error code: 2
17:29:07 INFO ~ 2023-10-27 17:29:07.072093: W tensorflow/stream_executor/gpu/asm_compiler.cc:80] Couldn't get ptxas version string: INTERNAL: Couldn't invoke ptxas.exe --version
17:29:07 INFO ~ 2023-10-27 17:29:07.076140: E tensorflow/core/platform/windows/subprocess.cc:287] Call to CreateProcess failed. Error code: 2
17:29:07 INFO ~ 2023-10-27 17:29:07.076637: W tensorflow/stream_executor/gpu/redzone_allocator.cc:314] INTERNAL: Failed to launch ptxas
17:29:07 INFO ~ Relying on driver to perform ptx compilation.
17:29:07 INFO ~ Modify $PATH to customize ptxas location.
17:29:07 INFO ~ This message will be only logged once.
17:29:10 INFO ~ Recent segmentation saved to: C:\Users\feige\AppData\Local\Temp\tmpacltb2wu\segmentation_segmentation
17:29:10 INFO ~ Finished DeepCell-predict
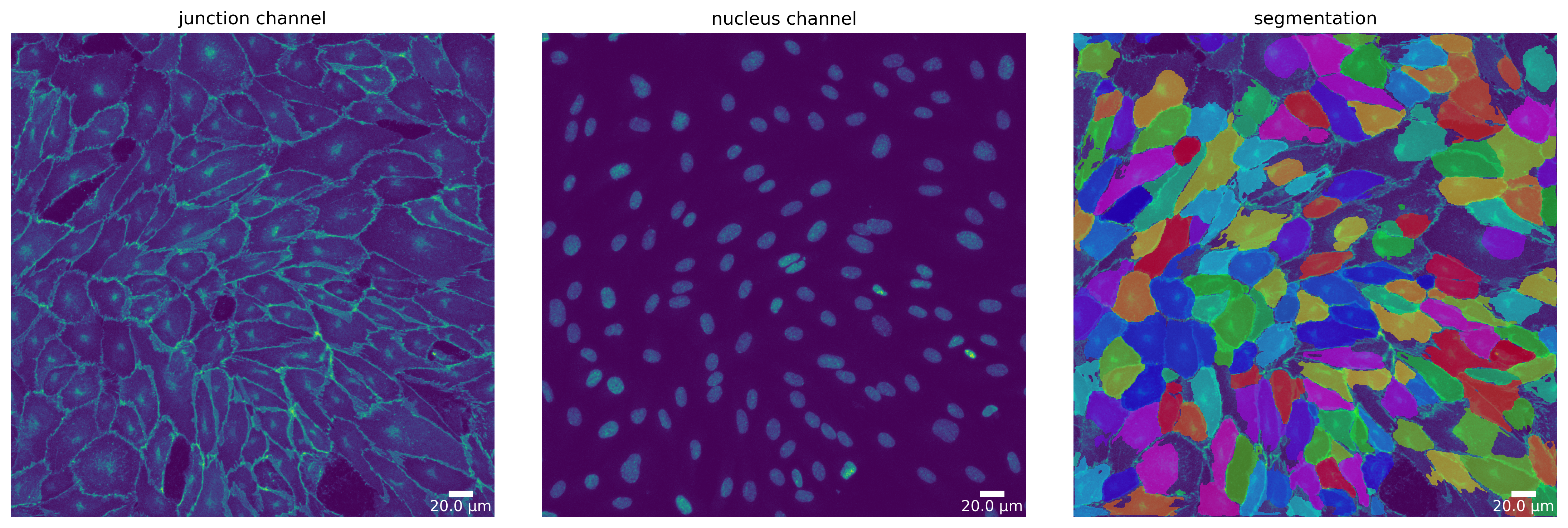
[15]:
# microSam
ms_mask = ms_seg.segment(ms_seg_prep)
plotter.plot_mask(ms_mask, ms_seg_prep, ms_seg_prep_p, output_path, output_file_prefix);
17:29:25 INFO ~ Starting microSAM-predict
17:30:01 INFO ~
17:30:08 INFO ~ Predict masks for point grid prompts: 0%| | 0/16 [00:00<?, ?it/s]
17:30:15 INFO ~ Predict masks for point grid prompts: 6%|▋ | 1/16 [00:06<01:42, 6.80s/it]
17:30:21 INFO ~ Predict masks for point grid prompts: 12%|█▎ | 2/16 [00:13<01:32, 6.61s/it]
17:30:29 INFO ~ Predict masks for point grid prompts: 19%|█▉ | 3/16 [00:20<01:27, 6.69s/it]
17:30:36 INFO ~ Predict masks for point grid prompts: 25%|██▌ | 4/16 [00:27<01:25, 7.10s/it]
17:30:42 INFO ~ Predict masks for point grid prompts: 31%|███▏ | 5/16 [00:34<01:15, 6.90s/it]
17:30:48 INFO ~ Predict masks for point grid prompts: 38%|███▊ | 6/16 [00:40<01:06, 6.70s/it]
17:30:54 INFO ~ Predict masks for point grid prompts: 44%|████▍ | 7/16 [00:46<00:59, 6.58s/it]
17:31:01 INFO ~ Predict masks for point grid prompts: 50%|█████ | 8/16 [00:53<00:51, 6.47s/it]
17:31:07 INFO ~ Predict masks for point grid prompts: 56%|█████▋ | 9/16 [00:59<00:45, 6.45s/it]
17:31:14 INFO ~ Predict masks for point grid prompts: 62%|██████▎ | 10/16 [01:06<00:38, 6.43s/it]
17:31:20 INFO ~ Predict masks for point grid prompts: 69%|██████▉ | 11/16 [01:12<00:31, 6.37s/it]
17:31:26 INFO ~ Predict masks for point grid prompts: 75%|███████▌ | 12/16 [01:18<00:25, 6.34s/it]
17:31:33 INFO ~ Predict masks for point grid prompts: 81%|████████▏ | 13/16 [01:24<00:19, 6.36s/it]
17:31:39 INFO ~ Predict masks for point grid prompts: 88%|████████▊ | 14/16 [01:31<00:12, 6.43s/it]
17:31:46 INFO ~ Predict masks for point grid prompts: 94%|█████████▍| 15/16 [01:37<00:06, 6.44s/it]
17:31:46 INFO ~ Predict masks for point grid prompts: 100%|██████████| 16/16 [01:44<00:00, 6.58s/it]
17:31:46 INFO ~ Predict masks for point grid prompts: 100%|██████████| 16/16 [01:44<00:00, 6.55s/it]
17:31:58 INFO ~ Finished microSAM-predict
Segmentation Mode “Nuclei”
Each segmentation algorithm supported in Polarity-JaM has several modi for different segmentation tasks. For example, Cellpose can segment nuclei and cytoplasm (based on the junction channel).
Here, we show how to switch to the “nucleus” mode for all three segmentation algorithms and compare the results.
[16]:
# Cellpose - switch to "nucleus" mode
params_runtime.segmentation_algorithm = "CellposeSegmenter"
cp_seg, _ = load_segmenter(params_runtime, cp_seg_p)
cp_mask_nuc = cp_seg.segment(cp_seg_prep, mode=SegmentationMode.NUCLEUS)
plotter.plot_mask(cp_mask_nuc, cp_seg_prep, cp_seg_prep_p, output_path, output_file_prefix);
cellprob_threshold and dist_threshold are being deprecated in a future release, use mask_threshold instead
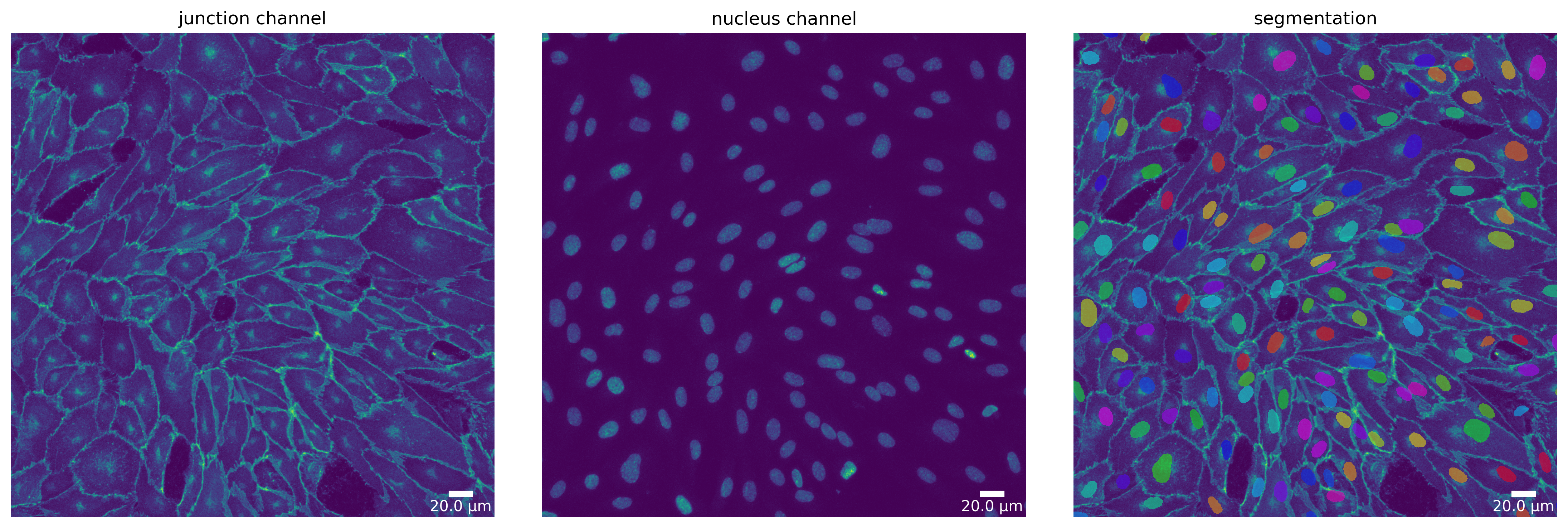
[17]:
# Deepcell - switch to "nucleus" mode
params_runtime.segmentation_algorithm = "DeepCellSegmenter"
dc_seg, _ = load_segmenter(params_runtime)
dc_mask_nuc = dc_seg.segment(dc_seg_prep, mode=SegmentationMode.NUCLEUS)
plotter.plot_mask(dc_mask_nuc, dc_seg_prep, dc_seg_prep_p, output_path, output_file_prefix);
17:32:31 INFO
Solution credits:
Noah F. Greenwald and Geneva Miller and Erick Moen and Alex Kong and Adam Kagel and Thomas Dougherty and Christine Camacho Fullaway and Brianna J. McIntosh and Ke Xuan Leow and Morgan Sarah Schwartz and Cole Pavelchek and Sunny Cui and Isabella Camplisson and Omer Bar-Tal and Jaiveer Singh and Mara Fong and Gautam Chaudhry and Zion Abraham and Jackson Moseley and Shiri Warshawsky and Erin Soon and Shirley Greenbaum and Tyler Risom and Travis Hollmann and Sean C. Bendall and Leeat Keren and William Graf and Michael Angelo and David Van Valen; Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning (DOI: https://doi.org/10.1038/s41587-021-01094-0)
17:32:33 INFO ~ Starting DeepCell-predict
17:32:38 INFO ~ 2023-10-27 17:32:38.795398: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX AVX2
17:32:38 INFO ~ To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
17:32:39 INFO ~ 2023-10-27 17:32:39.311336: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 21662 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3090, pci bus id: 0000:80:00.0, compute capability: 8.6
17:32:47 INFO ~ WARNING:tensorflow:No training configuration found in save file, so the model was *not* compiled. Compile it manually.
17:32:47 INFO ~ Image resolution the network was trained on: 0.5 microns per pixel
17:32:51 INFO ~ 2023-10-27 17:32:51.516871: I tensorflow/stream_executor/cuda/cuda_dnn.cc:368] Loaded cuDNN version 8800
17:32:52 INFO ~ 2023-10-27 17:32:52.569061: E tensorflow/core/platform/windows/subprocess.cc:287] Call to CreateProcess failed. Error code: 2
17:32:52 INFO ~ 2023-10-27 17:32:52.570483: E tensorflow/core/platform/windows/subprocess.cc:287] Call to CreateProcess failed. Error code: 2
17:32:52 INFO ~ 2023-10-27 17:32:52.570611: W tensorflow/stream_executor/gpu/asm_compiler.cc:80] Couldn't get ptxas version string: INTERNAL: Couldn't invoke ptxas.exe --version
17:32:52 INFO ~ 2023-10-27 17:32:52.576128: E tensorflow/core/platform/windows/subprocess.cc:287] Call to CreateProcess failed. Error code: 2
17:32:52 INFO ~ 2023-10-27 17:32:52.576667: W tensorflow/stream_executor/gpu/redzone_allocator.cc:314] INTERNAL: Failed to launch ptxas
17:32:52 INFO ~ Relying on driver to perform ptx compilation.
17:32:52 INFO ~ Modify $PATH to customize ptxas location.
17:32:52 INFO ~ This message will be only logged once.
17:33:05 INFO ~ Recent segmentation saved to: C:\Users\feige\AppData\Local\Temp\tmplrlc2oo1\segmentation_segmentation
17:33:05 INFO ~ Finished DeepCell-predict
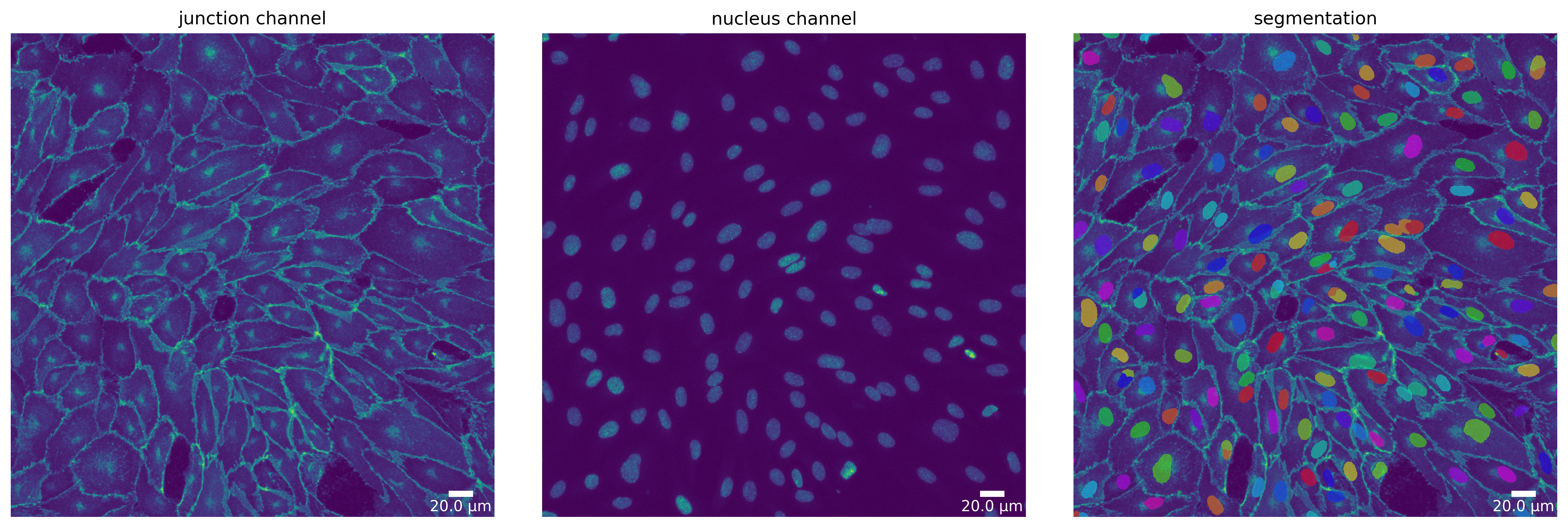
[18]:
# microSam - switch to "nucleus" mode
params_runtime.segmentation_algorithm = "MicrosamSegmenter"
ms_seg, _ = load_segmenter(params_runtime)
ms_mask_nuc = ms_seg.segment(ms_seg_prep, mode=SegmentationMode.NUCLEUS)
plotter.plot_mask(ms_mask_nuc, ms_seg_prep, ms_seg_prep_p, output_path, output_file_prefix);
17:33:18 INFO ~ Starting microSAM-predict
17:33:53 INFO ~
17:33:59 INFO ~ Predict masks for point grid prompts: 0%| | 0/16 [00:00<?, ?it/s]
17:34:06 INFO ~ Predict masks for point grid prompts: 6%|▋ | 1/16 [00:06<01:40, 6.67s/it]
17:34:12 INFO ~ Predict masks for point grid prompts: 12%|█▎ | 2/16 [00:13<01:30, 6.49s/it]
17:34:18 INFO ~ Predict masks for point grid prompts: 19%|█▉ | 3/16 [00:19<01:23, 6.42s/it]
17:34:25 INFO ~ Predict masks for point grid prompts: 25%|██▌ | 4/16 [00:25<01:17, 6.46s/it]
17:34:32 INFO ~ Predict masks for point grid prompts: 31%|███▏ | 5/16 [00:32<01:12, 6.56s/it]
17:34:38 INFO ~ Predict masks for point grid prompts: 38%|███▊ | 6/16 [00:39<01:06, 6.63s/it]
17:34:45 INFO ~ Predict masks for point grid prompts: 44%|████▍ | 7/16 [00:45<00:59, 6.56s/it]
17:34:51 INFO ~ Predict masks for point grid prompts: 50%|█████ | 8/16 [00:52<00:52, 6.53s/it]
17:34:58 INFO ~ Predict masks for point grid prompts: 56%|█████▋ | 9/16 [00:58<00:45, 6.48s/it]
17:35:04 INFO ~ Predict masks for point grid prompts: 62%|██████▎ | 10/16 [01:05<00:38, 6.45s/it]
17:35:10 INFO ~ Predict masks for point grid prompts: 69%|██████▉ | 11/16 [01:11<00:32, 6.46s/it]
17:35:17 INFO ~ Predict masks for point grid prompts: 75%|███████▌ | 12/16 [01:17<00:25, 6.44s/it]
17:35:23 INFO ~ Predict masks for point grid prompts: 81%|████████▏ | 13/16 [01:24<00:19, 6.40s/it]
17:35:30 INFO ~ Predict masks for point grid prompts: 88%|████████▊ | 14/16 [01:30<00:12, 6.38s/it]
17:35:36 INFO ~ Predict masks for point grid prompts: 94%|█████████▍| 15/16 [01:37<00:06, 6.43s/it]
17:35:36 INFO ~ Predict masks for point grid prompts: 100%|██████████| 16/16 [01:43<00:00, 6.48s/it]
17:35:36 INFO ~ Predict masks for point grid prompts: 100%|██████████| 16/16 [01:43<00:00, 6.48s/it]
17:35:49 INFO ~ Finished microSAM-predict
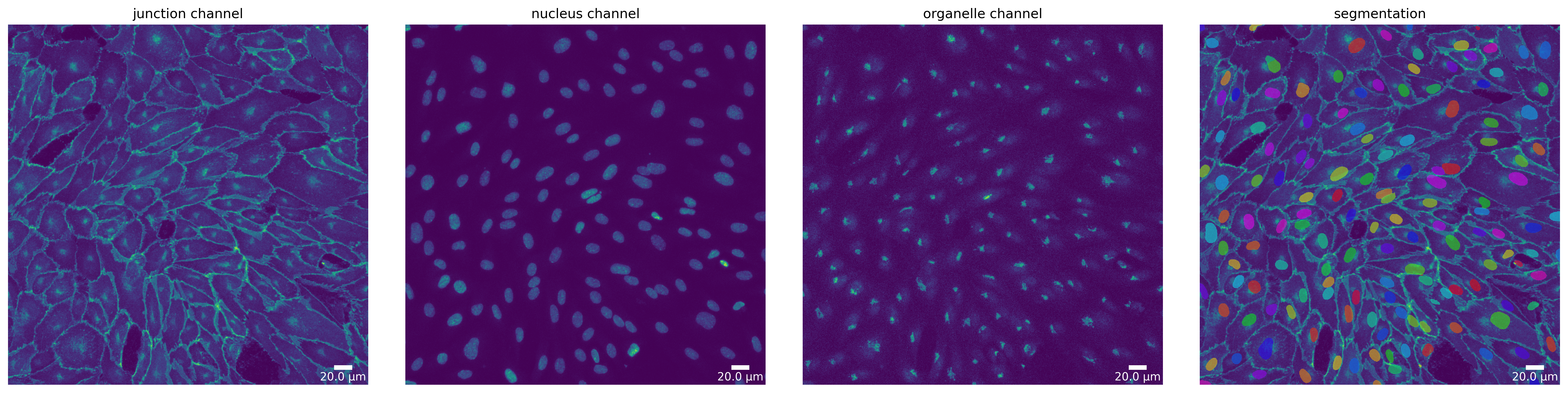
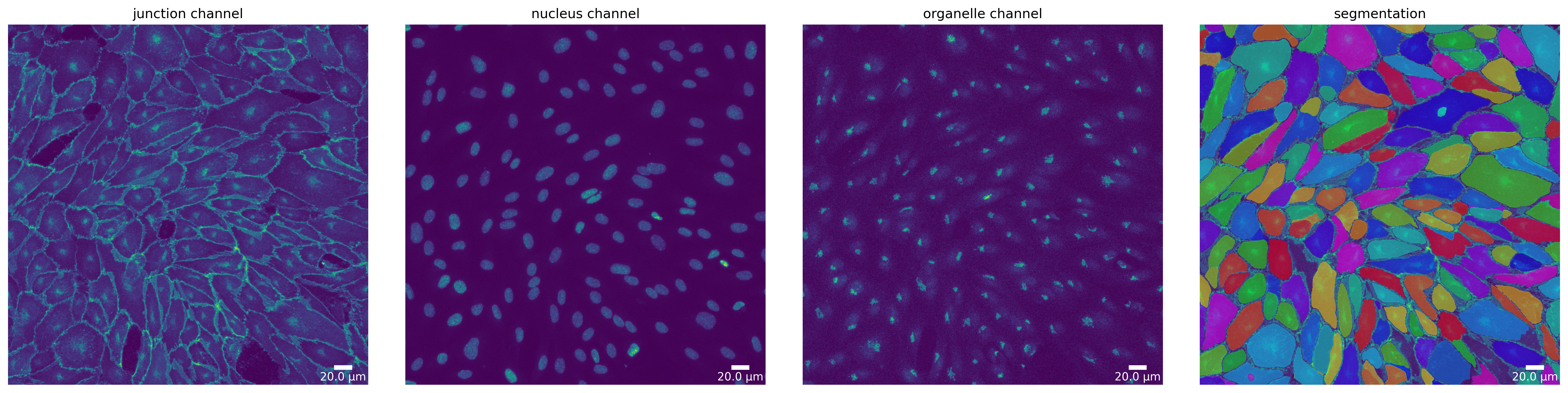
Segmentation Mode “Organelle”
Currently, only microSam supports the “organelle” segmentation mode. As the models focus is not the segmentation of nuclei, results should be taken with a grain of salt. Nevertheless, here we show how to use it.
[19]:
# microSam - switch to "organelle" mode
params_runtime.segmentation_algorithm = "MicrosamSegmenter"
# alter default parameters to enhance segmentation output
ms_seg_p = SegmentationParameter(params_runtime.segmentation_algorithm)
ms_seg_p.pred_iou_thresh = 0.85
# load segmenter with altered parameters
ms_seg, _ = load_segmenter(params_runtime, ms_seg_p)
ms_mask_nuc = ms_seg.segment(ms_seg_prep, mode=SegmentationMode.ORGANELLE)
plotter.plot_mask(ms_mask_nuc, ms_seg_prep, ms_seg_prep_p, output_path, output_file_prefix);
17:36:02 INFO ~ Starting microSAM-predict
17:36:37 INFO ~
17:36:43 INFO ~ Predict masks for point grid prompts: 0%| | 0/16 [00:00<?, ?it/s]
17:36:49 INFO ~ Predict masks for point grid prompts: 6%|▋ | 1/16 [00:05<01:29, 5.97s/it]
17:36:54 INFO ~ Predict masks for point grid prompts: 12%|█▎ | 2/16 [00:11<01:22, 5.86s/it]
17:37:00 INFO ~ Predict masks for point grid prompts: 19%|█▉ | 3/16 [00:17<01:14, 5.71s/it]
17:37:06 INFO ~ Predict masks for point grid prompts: 25%|██▌ | 4/16 [00:23<01:08, 5.73s/it]
17:37:12 INFO ~ Predict masks for point grid prompts: 31%|███▏ | 5/16 [00:28<01:03, 5.77s/it]
17:37:17 INFO ~ Predict masks for point grid prompts: 38%|███▊ | 6/16 [00:34<00:57, 5.75s/it]
17:37:23 INFO ~ Predict masks for point grid prompts: 44%|████▍ | 7/16 [00:40<00:51, 5.70s/it]
17:37:29 INFO ~ Predict masks for point grid prompts: 50%|█████ | 8/16 [00:45<00:45, 5.72s/it]
17:37:35 INFO ~ Predict masks for point grid prompts: 56%|█████▋ | 9/16 [00:51<00:40, 5.76s/it]
17:37:40 INFO ~ Predict masks for point grid prompts: 62%|██████▎ | 10/16 [00:57<00:34, 5.74s/it]
17:37:46 INFO ~ Predict masks for point grid prompts: 69%|██████▉ | 11/16 [01:03<00:28, 5.78s/it]
17:37:52 INFO ~ Predict masks for point grid prompts: 75%|███████▌ | 12/16 [01:09<00:23, 5.81s/it]
17:37:58 INFO ~ Predict masks for point grid prompts: 81%|████████▏ | 13/16 [01:15<00:17, 5.86s/it]
17:38:04 INFO ~ Predict masks for point grid prompts: 88%|████████▊ | 14/16 [01:21<00:11, 5.85s/it]
17:38:10 INFO ~ Predict masks for point grid prompts: 94%|█████████▍| 15/16 [01:26<00:05, 5.83s/it]
17:38:10 INFO ~ Predict masks for point grid prompts: 100%|██████████| 16/16 [01:32<00:00, 5.89s/it]
17:38:10 INFO ~ Predict masks for point grid prompts: 100%|██████████| 16/16 [01:32<00:00, 5.81s/it]
17:38:15 INFO ~ Finished microSAM-predict